
This increased usage has resulted in a real need for improved information retrieval (IR) methods. To manage the rapidly increasing amount of big, complex data, database information systems are increasingly leveraged ( Stein 2010, Lange et al. In recent years, information-processing methods have evolved from library research and individual data archives to web-based systems, cloud-computing and distributed databases. 2014).ĭespite this enormous amount of publicly available information, the search for candidate genes and relevant genomic data is a time-consuming and sophisticated task ( Divoli et al. Overall, >1,552 life science databases are publicly available ( Fernández-Suárez et al. 2011) or RAP-DB, the Rice Annotation Project Database ( Sakai et al. Furthermore, a number of individual platforms for different organisms, such as the Arabidopsis information resource (TAIR Lamesch et al. 2013) are two ontology information systems. Many information systems are specified in different broad subareas for example, Gene Ontology (GO Ashburner et al. Additionally, the NCBI GenBank plant division provides access to around 25 million sequences ( NCBI Nucleotide Plant Division Statistics 2014, ) and PubMed comprises >24 million citations for biomedical literature from MEDLINE, life science journals and online books ( NCBI Nucleotide PubMed Statistics 2014, ). In September 2014, the UniProt protein knowledge base had over 82.6 million entries ( UniProt Release Statistics 2014, ).
As a consequence, the number of annotated and functionally analyzed plant genomes, and publications of these genomes and gene products, is also on the rise. With the current wave of new and cheap technologies, vast amounts of data are being generated at an unprecedented rate ( Schadt et al.
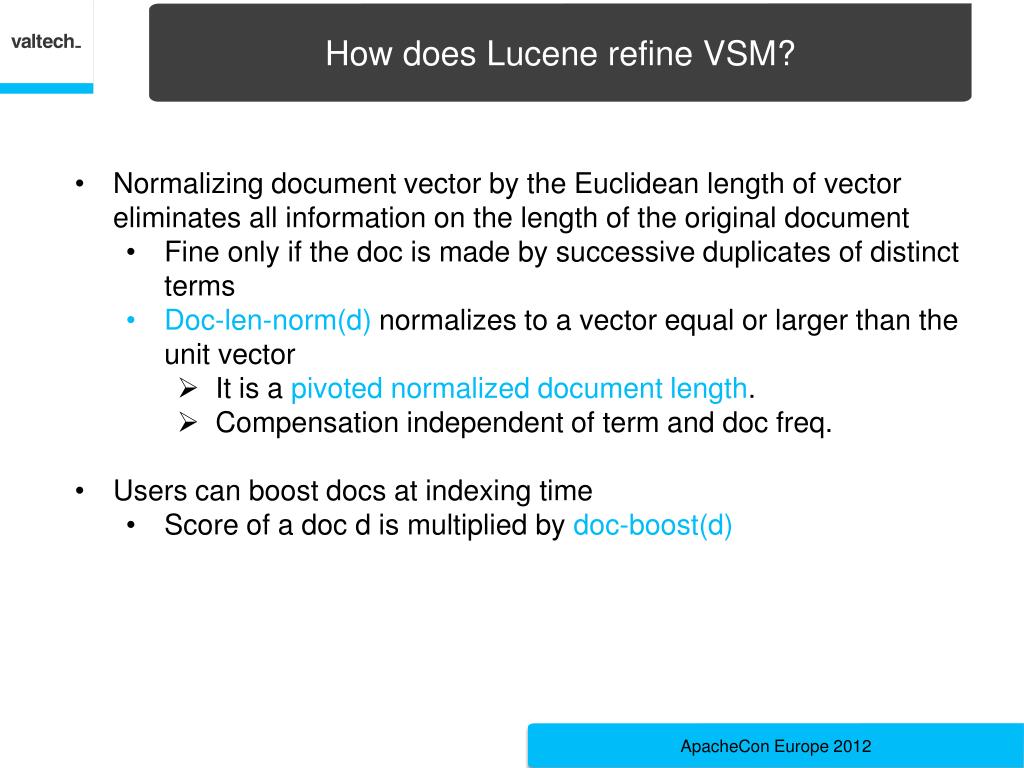
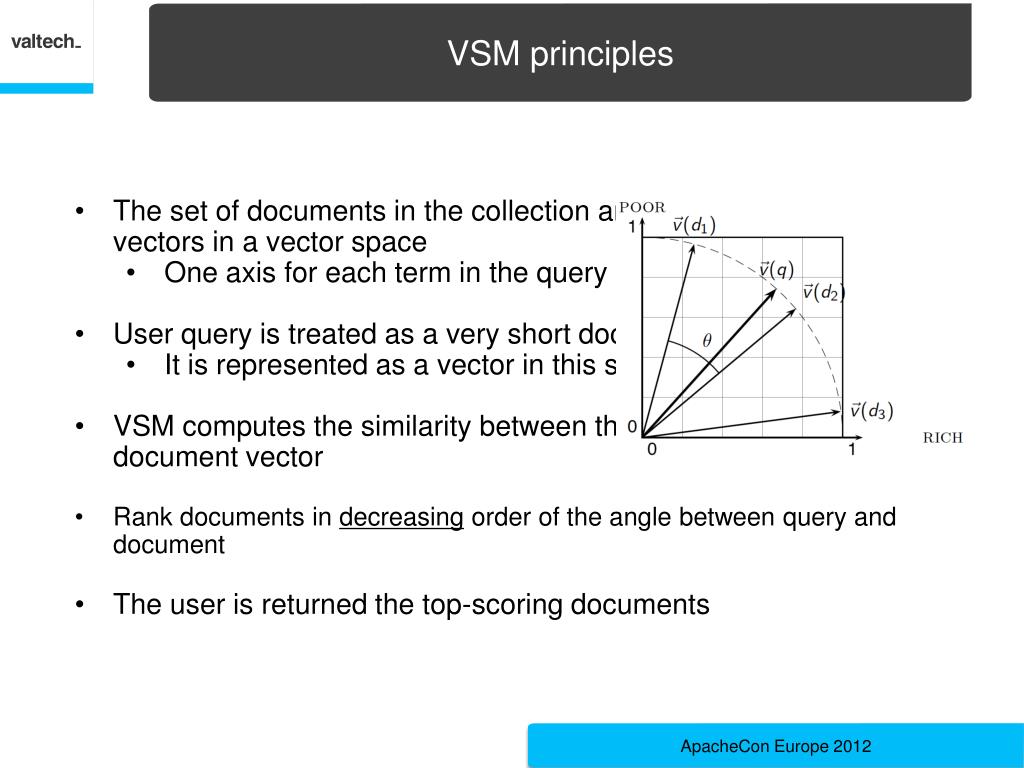
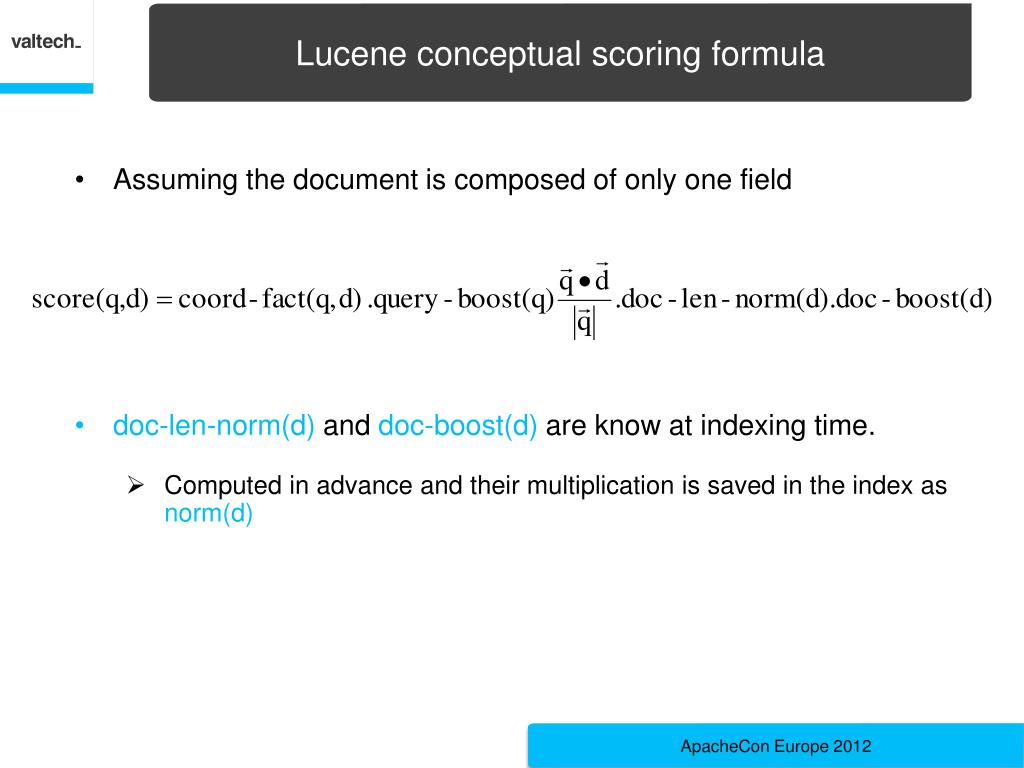
Modern molecular biology encompasses a broad range of methodologies, ranging from experimental data acquisition on genes and proteins to post-genomics technologies, such as RNA sequencing, phenotyping, proteomics, systems biology and integrative bioinformatics ( Kitano 2002). We fully describe LAILAPS’s functionality and capabilities by comparing this system’s performance with other widely used systems and by reporting both a validation in maize and a knowledge discovery use-case focusing on candidate genes in barley. An artificial neural network incorporating user feedback and behavior tracking allows relevance sorting of results. Query assistance and an evidence-based annotation system enable time-efficient and comprehensive information retrieval. The LAILAPS search engine allows fuzzy querying for candidate genes linked to specific traits over a loosely integrated system of indexed and interlinked genome databases. LAILAPS comprises around 65 million indexed documents, encompassing >13 major life science databases with around 80 million links to plant genomic resources. Here we describe LAILAPS ( ), an IR system designed to link plant genomic data in the context of phenotypic attributes for a detailed forward genetic research. Information retrieval (IR) has become an all-encompassing bioinformatics methodology for extracting knowledge from complex, heterogeneous and distributed databases, and therefore can be a useful tool for obtaining a comprehensive view of plant genomics, from genes to traits. Unfortunately, the information available today is widely scattered over a number of different databases. The association between genes and phenotypic traits is currently of great interest.

With the number of sequenced plant genomes growing, the number of predicted genes and functional annotations is also increasing.


 0 kommentar(er)
0 kommentar(er)
